Blog
P2 Allele of MYOT
Introduction
This blog post explores the P2 allele (S232P) of the equine MYOT gene, which encodes myotilin. Portions of this blog post serve as additional sources of information to supplement the MYOT Gene Page.
We present data to support the hypothesis that the P2 allele of MYOT (S232P) is damaging.
The substitution of proline (heterocyclic) for serine (polar uncharged) in the P2 variant of MYOT is a nonconservative substitution of a chemically dissimilar amino acid.
Evolutionary conservation provides convincing evidence that the P2 allele of MYOT is damaging. We use public data to show that the reference allele is widely conserved across most of 244 species of mammals and birds covering over 310 million years of evolutionary history.
There is a prominent exception in humans, where the MYOT gene has a conservative amino acid substitution that has gone to fixation (S232T). The substitution of threonine (polar uncharged) for serine (polar uncharged) in human MYOT is a conservative substitution of a chemically similar amino acid. This variant has gone to fixation in all the great apes most closely related to humans (chimp, bonobo, gorilla, and orangutan), but is absent from Old World monkeys, placing the origin in the common ancestor to great apes that diverged from other primates.
Correction of the MYOT protein model
We first offer a correction to public data from NCBI. The best protein model of equine MYOT is XP_014586147.2. This protein model carries the P2 allele of MYOT (S232P), which disagrees with the equine genomic sequence EquCab3.0, as shown in Figure 1.

Figure 1. A view of the UCSC browser centered on the position of the P2 variant at chr14:37,818,823 A/G. The coding sequence is on the reverse strand, so this sequence should be read right to left. The serine codon in the second frame of the three-frame translation at the top of the image is outlined in green, as is the S232 residue in the third protein model. Position 232 is a TCT codon, encoding serine. The P2 variant changes the TCT codon (serine) to CCT (proline).
Part of the sequence of equine MYOT protein model XP_014586147.2 is shown below, aligned with protein sequence predicted from genomic sequence EquCab3.0. The amino acid affected by the P2 allele is highlighted in red.
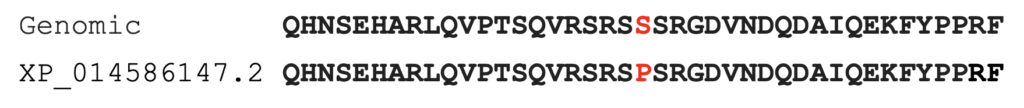
In the analysis presented below, the equine MYOT protein model XP_014586147.2 is corrected to the S232 allele present in the genomic sequence EquCab 3.0.
Evolutionary Conservation: Horse
Evolutionary conservation provides evidence on whether the P2 allele of MYOT is the derived allele. In this approach, predicted MYOT protein sequences are compared among a number of different species. This method, applied to species closely related to horse, is shown below.

Figure 2. Alignment of partial MYOT protein sequences from species closely related to horse. The horse sequence was corrected as described above, and used as a blastp query sequence to retrieve MYOT protein sequences from related species. CLUSTAL output summarizes whether a particular position is a single and fully conserved residue (*), has a conservative substitution with strongly similar properties (:), a somewhat conservative substitution (.), or is not conserved ( ). The position of S232 is highlighted in red. See the Technical Appendix for details.
Only the MYOT protein model from Przewalskii’s horse has the S232P allele. All other species closely related to horse (odd-toed ungulates) have a serine at this position. This argues strongly that the reference allele is S232, and the derived allele is P232.
CLUSTAL scores the S232P allele represented by the Przewalskii’s horse sequence as the absence of conservation at this position. We think that it is unlikely that MYOT-S232P has gone to fixation in this species.
Evolutionary Conservation: Human
The human MYOT protein (NP_006781.1) differs slightly from the corrected horse model as shown in the alignment of partial sequences below.
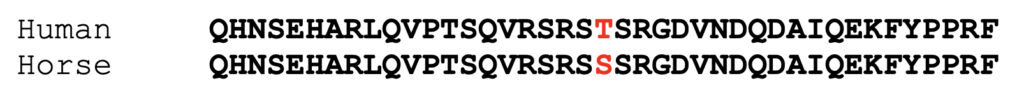
The human MYOT protein has a threonine at the position of the serine found in horse. This is a conservative substitution. Both serine and threonine have polar uncharged R groups and are phosphorylated in some proteins by a set of serine/threonine kinases.
Comparing the human MYOT protein sequence to species most closely related to humans (primates) gives the results shown below.

Figure 3. Alignment of partial MYOT protein sequences from species closely related to human. The human sequence was used as a blastp query sequence to retrieve MYOT protein sequences from related species. CLUSTAL output summarizes whether a particular position is a single and fully conserved residue (*), has a conservative substitution with strongly similar properties (:), a somewhat conservative substitution (.), or is not conserved ( ). The position of S232 is highlighted in red. See the Technical Appendix for details.
Humans and the closely-related great apes (chimpanzee, bonobo, gorilla, and orangutan) have a threonine at position 232. Old World monkeys (snub-nosed monkey, gibbons, siamang, macaque, colobus, mangabey, baboon, leaf monkey, green monkey) and prosimians (tarsier and lemur) have a serine at position 232.
The substitution of threonine for serine at position 232 occurred in the common ancestor of great apes, branching off from other Old World Monkeys an estimated 18 million years ago.
Evolutionary Conservation: Mammals
Both horses and humans are mammals. We searched for partial MYOT protein sequences among a wide range of mammals. The human sequence was used as a blastp query sequence to retrieve MYOT protein sequences from mammals. Identical sequences were clustered. If a particular retrieved sequence was unique, it was also used as a blastp query sequence to recover additional sequences. Some sequences remained unique. The results are shown below.

Figure 4. Alignment of partial MYOT protein sequences from 108 mammals. Sequences that were identical were clustered. Numbers in parentheses indicate the number of species in a cluster. CLUSTAL output summarizes whether a particular position is a single and fully conserved residue (*), has a conservative substitution with strongly similar properties (:), a somewhat conservative substitution (.), or is not conserved ( ). The position of S232 is highlighted in red. See the Technical Appendix for details.
The alignment shown in Figure 4 demonstrates that the S232 allele is invariant throughout the mammalian lineage, with the exception of humans and the great apes. Conservation of the S232 allele is seen throughout mammals, including marsupials (wombat, koala, and some marsupials included in the mammalian clusters) and monotremes (platypus, echidna).
Evolutionary Conservation: Birds
The common ancestor of birds diverged from the common ancestor of mammals about 310 million years ago. We searched for partial MYOT protein sequences among a wide range of birds. The human sequence was used as a blastp query sequence to retrieve MYOT protein sequences from birds. Identical sequences were clustered. The first amino acid in the human sequence varies even among mammals, and is also not conserved among birds, with some species having an in-frame deletion that removes two amino acids at this position. The first amino acid was removed from all bird sequences in order to reduce the number of clusters.

Figure 5. Alignment of partial MYOT protein sequences from 136 birds. Identical sequences were clustered. Numbers in parentheses indicate the number of species in a cluster. CLUSTAL output summarizes whether a particular position is a single and fully conserved residue (*), has a conservative substitution with strongly similar properties (:), a somewhat conservative substitution (.), or is not conserved ( ). The position of S232 is highlighted in red. See the Technical Appendix for details.
In the alignment shown in Figure 5, only 22 of the 40 positions are invariant throughout the avian lineage; the S232 allele is invariant. This shows that the S232P allele is not tolerated over evolutionary time. The only tolerated substitution at this position is the S232T allele that has gone to fixation in humans and the great apes.
Summary
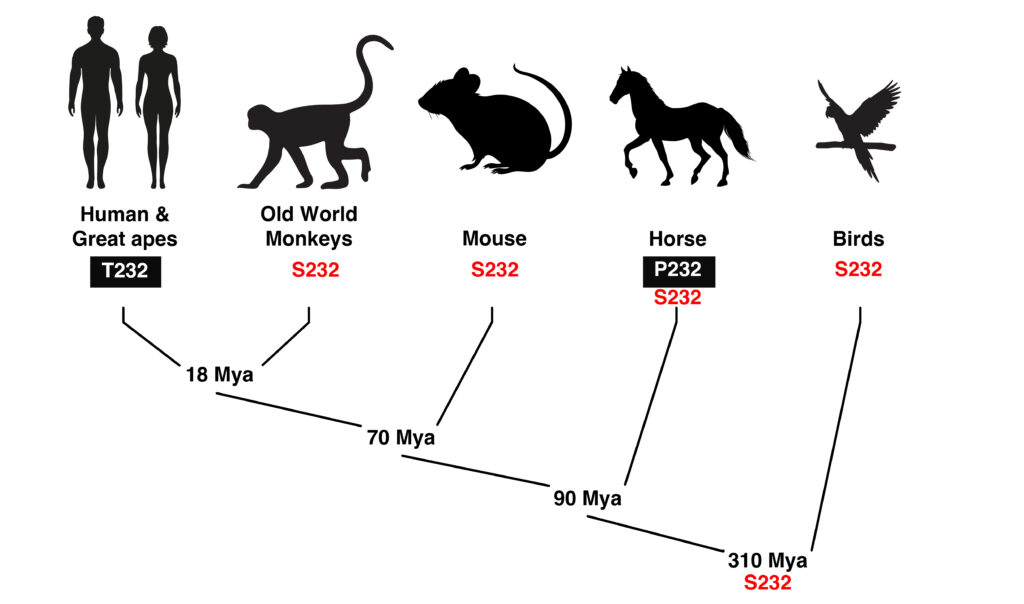
Figure 6. The amino acid at the position of the equine P2 allele of MYOT (S232P) is a serine conserved throughout evolution with two notable exceptions. First, the amino acid at this position in humans and great apes is the conservative substitution threonine (S232T), which appears to have gone to fixation when this lineage diverged from Old World monkeys. Second, the P2 allele of MYOT (S232P) appears as the minor allele in horse, and also in the closely related Przewalskii’s horse, but species closely related to horse have the ancestral S232, as do all birds. Estimated time of divergence is shown as millions of years ago (Mya).
Technical Appendix
The purpose of this technical appendix is to permit researchers to reproduce or extend these results independently.
UCSC Genome Browser. Here is a link to the UCSC Genome Browser centered on the base altered by the P2 allele of MYOT.
Retrieving protein sequences. Protein sequences like those shown in the alignments (Figures 2, 3, 4, and 5) can be retrieved from NCBI using the blastp tool and a query sequence.
The partial human MYOT query sequence used to retrieve sequences is:

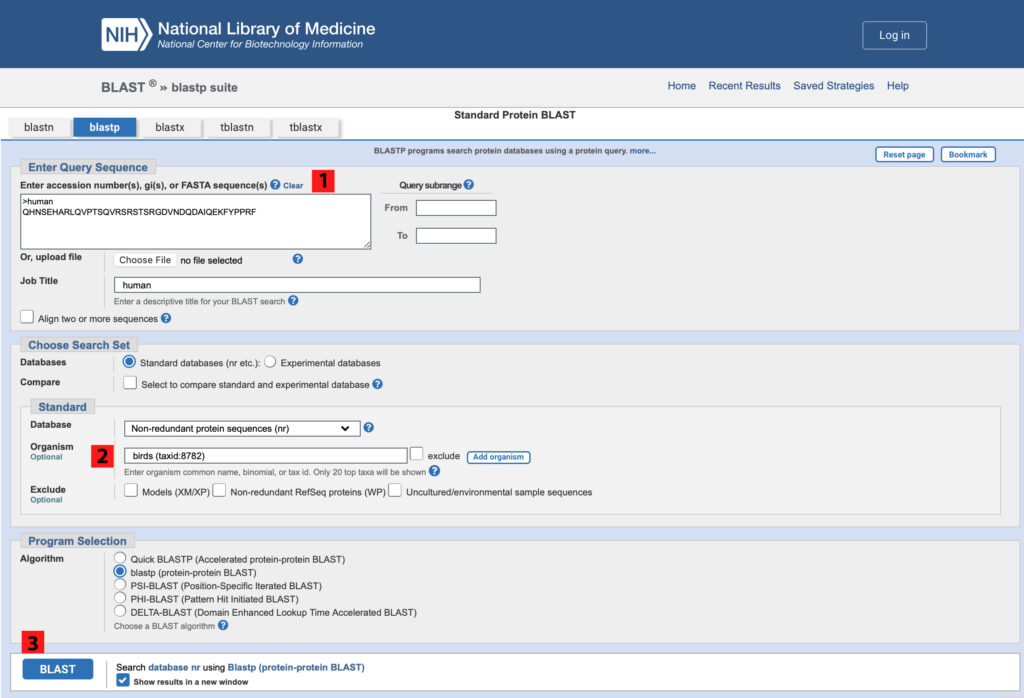
Figure 7. NCBI blastp search page. To use the human MYOT query sequence to identify MYOT sequences from birds, 1) copy the human sequence to the query sequence box, 2) set the optional Organism to “birds.” Note that this recovers a taxonomic ID as shown, and 3) Click BLAST.
Results for one species are shown below.

The match is to the Northern carmine bee-eater (Merops nubicus). The Query line shows the part of the human sequence that matched the bird sequence. The Subject line shows the bird sequence. At each position, if the amino acid is identical in the two sequences, the amino acid is entered in the middle line. Conservative substitutions are marked with a “+” sign; nonconservative substitutions are left blank.
The recovered sequence can be put into FASTA format:

This can then be used as a query sequence for an additional blastp search.
Aligning protein sequences
Multiple protein sequences were aligned using CLUSTAL.
Evolutionary relationships
Information on evolutionary relationships among species is presented graphically as the Tree of Life.
Download data
The data used for alignments in Figures 2, 3, 4, and 5 are available as a spreadsheet.
For each sequence used in the analysis, the spreadsheet contains the figure in which the sequence appears. Each individual species in a cluster is identified by species name and common name. The sequence ID for the MYOT protein sequence in that species is also shown.
Share this post
From the blog
The latest industry news, interviews, technologies, and resources.
Experienced Equestrian Joins EquiSeq Board
Albuquerque, New Mexico, USA June 20, 2025 EquiSeq, a leader in developing genetic…
