Blog
Genes First!
EquiSeq has released a major revision to the software that powers our website and database. We now present genetic test results starting with the affected gene. Here is an example of the appearance of the new horse profile pages.
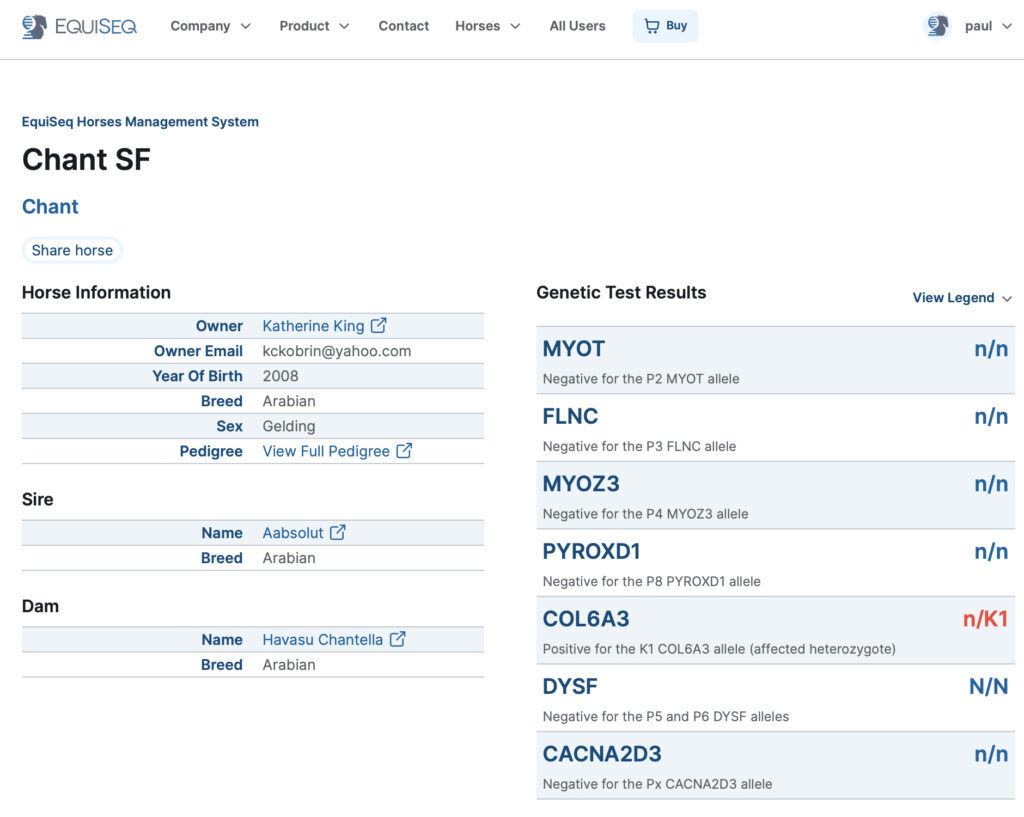
Figure 1. A horse profile page returning results from EquiSeq. This horse has tested positive for the K1 variant of COL6A3 (n/K1), negative for the other five variants in EquiSeq’s DNA test, and also negative for the P5 and P6 variants of DYSF, an experimental test not available commercially.
What is a gene?
In the context of mammalian genetics, a gene is a unit of hereditary information made of DNA. These segments of DNA encode proteins. Every mammal has two copies of every gene (except for X-linked genes in males). In a population, there are variant forms of genes, called alleles of that gene, that differ in their DNA sequence.
Some alleles produce proteins that differ from the reference or wild-type sequence in minor ways that do not affect protein function. Other alleles produce proteins that are nonfunctional or have an altered function, causing a change in the development or function of the organism.
What is a disease state?
Disease is defined as any deviation from the normal structural or functional state of an organism, generally associated with particular clinical signs and symptoms, and distinct from physical injury. Many disease states are progressive, with more clinical signs and symptoms appearing or worsening over time.
A particular disease state in horse can be recognized by specific clinical signs as interpreted by a veterinarian. This is the basis for a diagnosis, which can only be given by a veterinarian.
Example: Genetic variant associated with PSSM1
The disease state Polysaccharide Storage Myopathy, type 1 (PSSM1) is described by a set of clinical signs. The assay used to identify the genetic variant associated with this disease state is muscle biopsy. The presence of enlarged, amylase-resistant glycogen granules in muscle biopsy samples indicates PSSM1 (1). The association between this particular clinical sign and a genetic variant is strong enough to identify the variant in a genome-wide association study (GWAS). The genetic variant is a missense allele, which causes the substitution of one amino acid for another, in the gene encoding glycogen synthase 1 (GYS1). The full nomenclature for this variant, GYS1-R309H, gives the original amino acid, the position of this amino acid in the protein, and the new amino acid. The variant changes the amino acid at position 309 in the protein sequence from arginine (R) to histidine (H).
The altered glycogen synthase encoded by GYS1-R309H is constitutively activated. The activity of glycogen synthase is regulated by phosphorylation. Normally, glycogen synthase is fully activated when it is not phosphorylated, and relatively inactive when it is phosphorylated. The GYS1-R309H protein escapes this regulation and is fully active when phosphorylated (2).
The level of glycogen synthase activity is correlated with the genotype for GYS1. GYS1-R309H is commonly abbreviated P1. Horses that have two copies of the wild-type allele of GYS1 (n/n) have the lowest level of glycogen synthase activity, horses with two copies of the variant (P1/P1) have the highest levels, and horses heterozygous for the variant (n/P1) have intermediate levels (2).
Other clinical signs of PSSM1 are not perfectly correlated with the GYS1 genotype. Many horses that are n/P1 or P1/P1 do not show exercise intolerance, gait abnormalities, muscle wasting, or any other clinical signs associated with the disease state. Even the assay used to identify the variant, muscle biopsy, does not perfectly identify horses carrying the variant. Among Quarter Horses, 22.4% of horses scored as normal by muscle biopsy have one or two copies of the P1 variant, while 4.5% of horses scored as having enlarged, amylase-resistant glycogen granules do not carry the P1 variant (3).
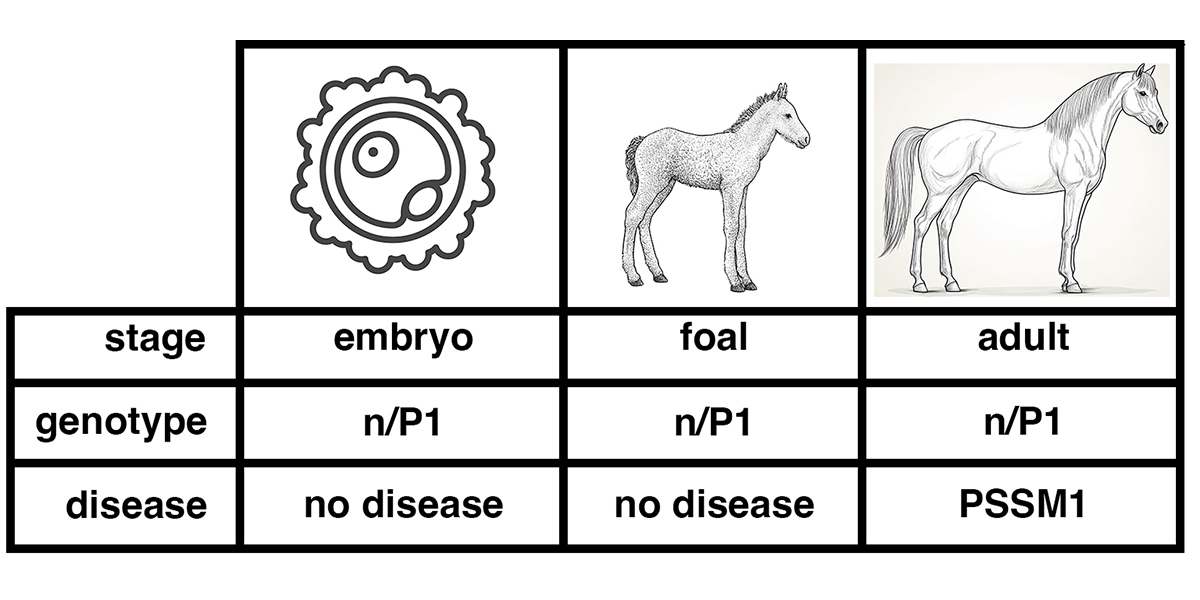
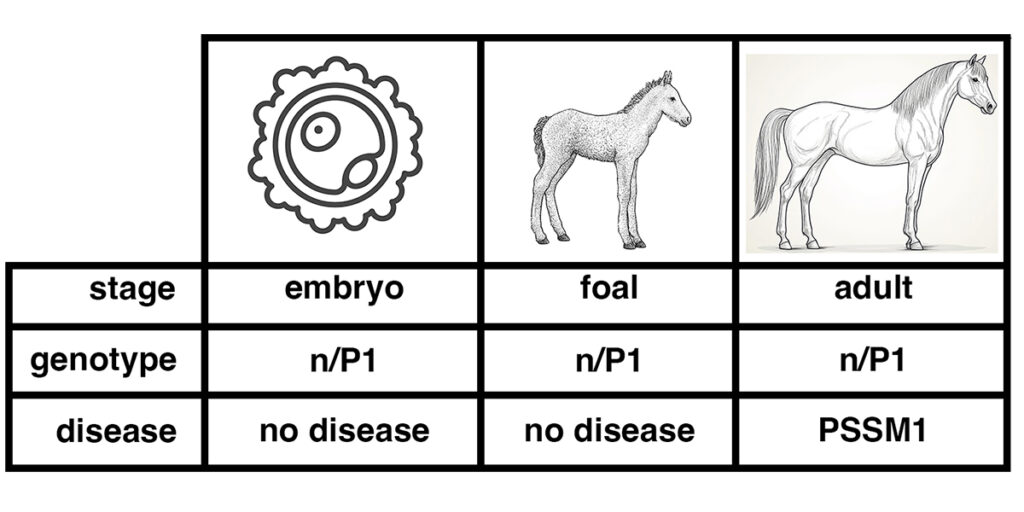
As shown in the figure below, the genotype of a horse – whether it is n/n, n/P1, or P1/P1 – does not change over the life of the horse. An individual cell taken from a pre-implantation embryo can be tested for P1. If this embryo develops to a foal, the foal will have the same genotype as the sampled embryonic cell. So will the adult horse.
In contrast, the disease state PSSM1 develops over time, influenced by environmental factors (diet and exercise) and possibly by other genetic variants.
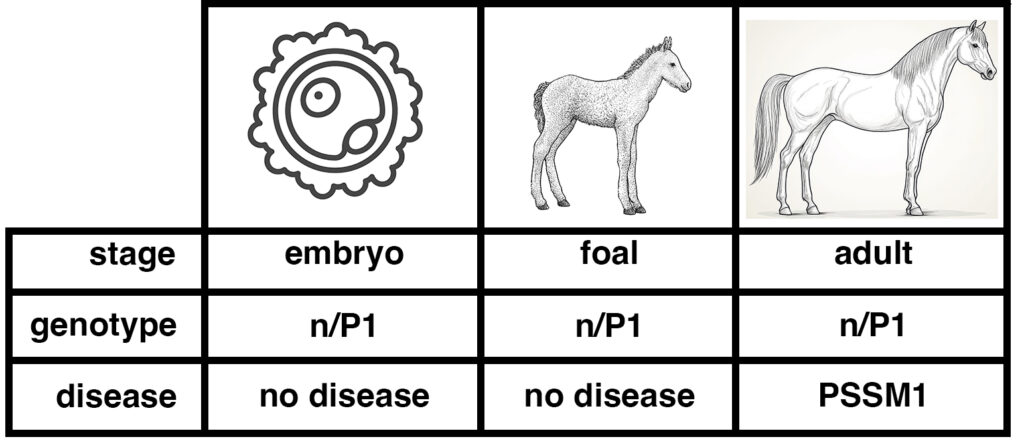
Figure 2. The genotype is set at conception, the disease state develops over time. DNA tests of a horse that is n/P1 will give the same result over the life of the horse, from embryo to foal to adult. Diagnosis of the disease state PSSM1 will usually only be made after the horse has reached adulthood.
Suppose an embryo cell tests positive for GYS1-R309H, with the genotype n/P1. The genotype indicates that the horse will have the potential to develop the disease state PSSM1.
Does this mean that the embryo has PSSM1? Of course not. The embryo at that stage lacks differentiated muscle tissue, so it cannot be said to have a muscle disease. A foal with the genotype n/P1 is unlikely to show symptoms of PSSM1. Many n/P1 adult horses show symptoms of PSSM1.
Why did EquiSeq change the way that it returns results?
We initially designed our website and database to return results in a form familiar to horse owners, following the example of results returned for AQHA testing through the University of California, Davis. Their nomenclature for results conflates the genotype with the disease state. Heterozygotes for GYS1-R309H are described as n/PSSM1.
We think that this is misleading. As discussed above, not all horses that are n/GYS1-R309H (n/P1) show enlarged, amylase-resistant glycogen granules on muscle biopsy, and some horses that do show this clinical sign do not carry GYS1-R309H (they are n/n). There may be genetic variants of other genes that account for horses that have enlarged, amylase-resistant glycogen granules. If a variant of a gene other than GYS1 is associated with a glycogen storage defect that looks like PSSM1, what would be the genetic nomenclature used? The UC Davis nomenclature only works if there is only one gene associated with a particular disease state, but even then, it conflates genetic testing results with a disease state.
In human genetics, it is common for genetic variants in different genes to produce a collection of symptoms that are clinically identical. For example, OMIM currently lists twelve different genes with variants that are associated with Myofibrillar Myopathy (MFM). Disease states in this case are named with different numbers, depending on the gene involved. The disease state MFM1 is associated with variants of desmin (DES), MFM2 is associated with variants of alpha-beta crystallin (CRYAB), and so on. Genotypes in these cases are given as homozygotes or heterozygotes for specific mutations; the abbreviation for the disease state is not used in genetic nomenclature.
As the number of genetic variants potentially associated with disease states in horse continues to increase, we think that the best solution is to report genetic test results in a way that is clear and unambiguous.
References
1. McCue, ME, et al. (2008) Glycogen synthase (GYS1) mutation causes a novel skeletal muscle glycogenosis. Genomics. 91(5):458-466. PubMed ID: 18358695
2. Maile, CA, et al. (2017) A highly prevalent equine glycogen storage disease is explained by constitutive activation of a mutant glycogen synthase. Biochim Biophys Acta Gen Subj. 1861(1 Pt A):3388-3398. PubMed ID: 27592162
3. McCue, ME et al. (2008) Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storage myopathy. J Vet Intern Med. 22(5):1228-1233. PubMed ID: 18691366
Share this post
From the blog
The latest industry news, interviews, technologies, and resources.
Twenty European countries issue patent on EquiSeq DNA tests
ALBUQUERQUE, NEW MEXICO EquiSeq announced today that a group of twenty countries in…