Blog
Genetic Basis of Exercise Intolerance in Arabians

Paul Szauter, PhD, Chief Scientific Officer of EquiSeq, gave a presentation titled “Genetic Basis of Exercise Intolerance in Arabians” at the Al Khamsa Annual Meeting and Convention in Fayetteville, Arkansas on October 13, 2019. EquiSeq is a company based in Albuquerque, New Mexico, that develops and sells genetic tests for horses.
Exercise intolerance is known in Arabians (1, 2) and limits the performance of some affected horses in endurance events. Symptoms of exercise intolerance (tying up) include sudden lameness, myoglobinuria, and greatly increased serum levels of creatine kinase (CK), a muscle enzyme released into the bloodstream when muscles are damaged.
Researchers from EquiSeq began studying exercise intolerance in Arabians at an endurance event at the Biltmore estate in 2017. They were stationed at the vet trailer in order to study horses pulled from the event for symptoms of lameness. The weather was unusually cold and rainy, and it wasn’t long before the first horse showed up at the vet trailer with urine the color of black coffee and obvious symptoms of pain and lameness. A test for serum CK (normally in the range of 500 or less) showed levels in excess of 20,000. Eight hours and thirty liters of IV saline later, the horse had recovered. The research team took a blood sample to isolate DNA for whole genome DNA sequencing.
There are several different types of exercise intolerance in horses, distinguished by symptoms and by findings on muscle biopsy (1). The incidence of the different types differs among breeds. Arabians are prone to a type called Arabian Myofibrillar Myopathy (MFM). In addition to the symptoms of lameness, myoglobinuria, and elevated serum CK, Arabian MFM shows a distinct finding on muscle biopsy. There is a disruption of the structure of the units of skeletal muscle called myofibrils. Normal muscle shows a very orderly pattern of repeating units of muscle tissue called sarcomeres. A sarcomere consists of overlapping thick (myosin) and thin (actin) filaments, with the thin filaments anchored to structures at either end of the sarcomere called Z discs. An inherited disorder called Myofibrillar Myopathy in human patients shows a disruption of the Z discs, resulting in disordered sarcomeres. Human patients experience a progressive weakening of their muscles. Human Myofibrillar Myopathy results from inherited defects in at least nine different genes; some cases cannot be assigned to known genes.
One of the clues about the genetic basis of exercise intolerance in Arabians comes from a study of gene and protein expression in Arabians with and without Arabian MFM (3). The study shows that affected Arabians have a defect in the defense against oxidative stress, specifically in cysteine metabolic pathways, resulting in a deficiency of cysteine-containing cellular antioxidants (such as peroxiredoxin). Aerobic exercise in these horses generates an excess of reactive oxygen species that damage muscle proteins.
All mammals are closely related, and the study of genetic disorders of muscle in humans and mice can guide the identification of the relevant genes in horses. Researchers at EquiSeq use whole genome sequencing of horses to identify all genetic variants in selected horses, picking out genetic variants observed in a list of candidate genes associated with inherited disorders of muscle in humans and mice. Each genetic variant in a candidate gene is evaluated by comparing the protein sequence encoded by the corresponding gene in many species of vertebrates, from humans to sharks. A mutation that changes a specific amino acid to a chemically different one can be evaluated by seeing whether that change has been seen in any other species over evolutionary time. Changes that are seen in other species in nature are scored as benign, while changes that are not seen in any other species are potentially pathogenic (disease-causing).
EquiSeq has used this approach to identify potentially pathogenic mutations in three different muscle proteins that are structural components of the Z disc: myotilin, filamin C, and myozenin 3. Mutations in the genes encoding these proteins (MYOT, FLNC, and MYOZ3) are associated with exercise intolerance in many breeds of horses, and are part of EquiSeq’s Myopathy Panel, a set of DNA tests to detect genetic defects that predispose to inherited exercise intolerance. These tests identify specific missense alleles of these three genes and are fully described on the EquiSeq website, although they have not yet been published in the peer-reviewed scientific literature. Defects in MYOT, FLNC, and MYOZ3 are not associated with myoglobinuria or greatly elevated serum CK, and are unlikely to be the basis of Arabian MFM.
The study of Arabians with exercise intolerance allowed researchers at EquiSeq to discover an additional genetic variant (nicknamed P8) in an undisclosed gene of known function. Defects in this gene are associated with Myofibrillar Myopathy in human patients, and the gene is expected to play a role in the defense against oxidative stress. Horses homozygous for the defect (P8/P8) show a pattern of muscle wasting similar to the group of human disorders called Limb-Girdle Muscular Dystrophy. Horses heterozygous for the defect (n/P8) also show symptoms of muscle wasting and exercise intolerance, especially in combination with mutations in MYOT or MYOZ3. In addition, it appears that both n/P8 and P8/P8 horses require higher levels of vitamin E supplementation to maintain normal serum vitamin E levels. Vitamin E is an antioxidant that protects lipids in plasma membranes from damage caused by reactive oxygen species. One molecule of vitamin E is consumed each time that a lipid peroxidation cycle is stopped.
Researchers at EquiSeq developed a DNA test that can be performed on a hair sample. This allowed the survey of 316 Arabians recruited from Arabian interest groups on Facebook without respect to symptoms to determine the incidence of the P8 allele, which is common in Arabians. The allele frequency is about 0.25, which means that the population of Arabians is about 6% P8/P8, 37% n/P8, and 57% n/n.
Researchers at EquiSeq collaborate with other research groups around the world. One group is focused on the biochemical characterization of the protein affected by the P8 genetic variant, and has been studying the effects of various mutations that cause amino acid substitutions in this protein. The amino acid substitution caused by the P8 genetic variant has been shown to be damaging, causing the protein to become unstable.
EquiSeq added the test for the P8 allele to its Myopathy Panel on the day of the presentation at the Al Khamsa Convention. All of the genetic variants in EquiSeq’s Myopathy Panel are currently under independent review in a public study being conducted at the University of Minnesota, funded in part by a grant from the Morris Animal Research Foundation. This test will allow breeders to breed out the P8 genetic variant, as shown in the figure. For many breeders, the primary focus of a breeding program for Al Khamsa Arabians is breed preservation, with an attempt to preserve as much genetic diversity as possible. A secondary goal should be the gradual elimination of genetic defects that affect performance, like the P8 genetic variant. In the case of critically endangered lines, it may be justifiable to breed a P8/P8 horse to a horse that is n/n. This will result in 100% n/P8 offspring. Ideally, in the next generation, these could be bred to n/n horses to give a 50% chance of n/n offspring.
Traditional criteria for breeding Al Khamsa Arabians, such as the preservation of bloodlines and the preservation of positive traits in appearance, temperament, and performance, can be combined with the emerging science of equine genomics through inexpensive DNA tests to eliminate genetic defects affecting performance in the Al Khamsa Arabians of the future.
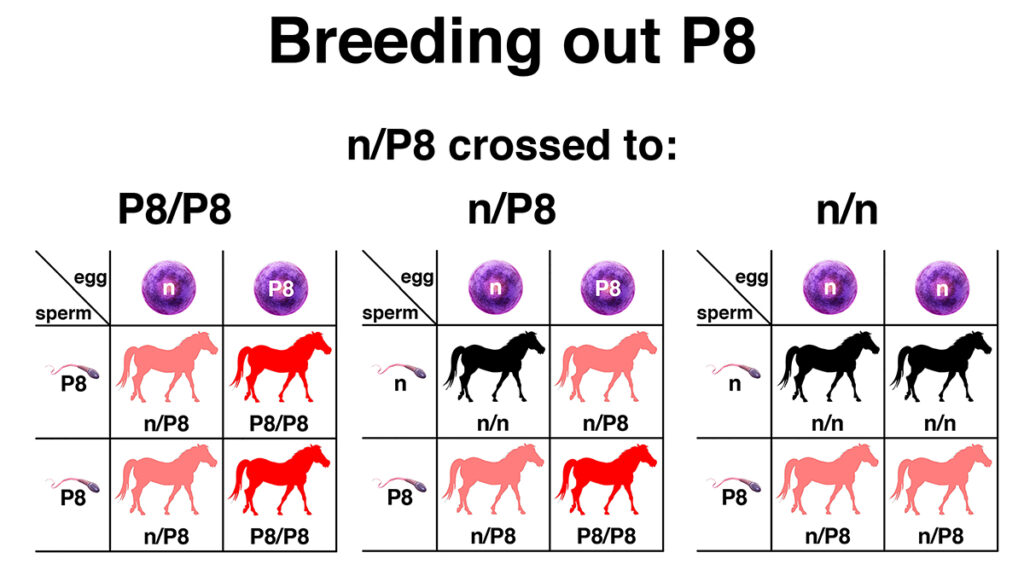
This figure shows three different Punnett squares that predict the outcomes of breeding a horse heterozygous for the P8 genetic variant (n/P8) to horses of each of the three possible genotypes (P8/P8, n/P8, and n/n). A foal from the cross of n/P8 to P8/P8 (left) has a 50% chance of being homozygous for P8 (P8/P8) and a 50% chance of being heterozygous for P8 (n/P8). A foal from the cross of n/P8 to n/P8 (center) has a 25% chance of being homozygous for P8 (P8/P8), a 50% chance of being heterozygous for P8 (n/P8), and a 25% chance of being free of the variant (n/n). A foal from the cross of n/P8 to n/n (right) has a 50% chance of being heterozygous (n/P8) and a 50% chance of being free of the variant (n/n). Genetic testing of hair samples will allow the P8 genetic variant to be bred out of Al Khamsa Arabians over several generations of selective breeding.
Please see the article on pages 18 and 19 in the online version of The Khamsat .
Citations
[1] Valberg, SJ (2018). “Muscle conditions affecting sport horses.” Vet Clin North Am Equine Pract. 34(2):253-276. PMID: 29853158.
[2] Valberg, SJ et al. (2016). “Suspected myofibrillar myopathy in Arabian horses with a history of exertional rhabdomyolysis.” Equine Vet J. 48(5):548-56. PMID: 26234161.
[3] Valberg, SJ et al. (2018). “Proteome and transcriptome profiling of equine myofibrillar myopathy identifies diminished peroxiredoxin 6 and altered cysteine metabolic pathways.” Physiol Genomics. 50 (12): 1036-1050. PMID: 30289745.
Share this post
From the blog
The latest industry news, interviews, technologies, and resources.
