Our blog
Resources and insights
The latest industry news, interviews, technologies, and resources.
Blog categories
Blog
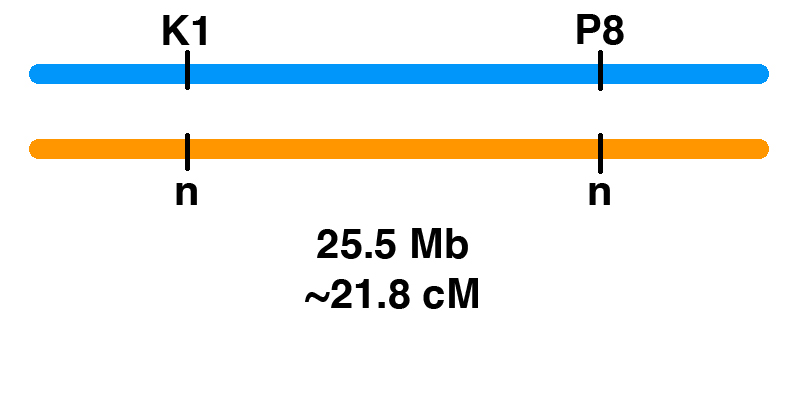
Genetic Linkage of P8 and K1
How can you figure out the chances of different genotypes when breeding horses?Monohybrid CrossIt’s easy for one gene. Let’s say that there is a stallion that has one copy of one of the genetic variants associated with PSSM2, the P2 variant (MYOT-S232P). This stallion is heterozygous (n/P2), meaning that he has one normal copy of the MYOT gene and one copy with the P2 mutation.If the stallion is bred to a mare that is clear (n/n) for the P2 allele, the Punnett square below shows the odds.Gametes (sperm and eggs) contain a single copy of each gene in the horse. The stallion produces two kinds of sperm in equal frequency: sperm with P2, and sperm with the normal allele (n). The mare produces eggs with one of the two normal alleles (n).This means that there is a 50% chance of a foal that is n/P2, and a 50% chance of a foal that is n/n.Dihybrid CrossWhat if we are following two genes? In general, two gene pairs will show independent assortment. This means that the segregation of one pair of alleles into gametes will not influence the segregation of another pair of alleles.If we tossed two coins at the same…
News
Generatio GmbH – Center for Animal Genetics Adds P8 and K1 Tests
Tubingen, Germany Generatio GmbH – Center for Animal Genetics (CAG) has added the P8 variant of PYROXD1 and the K1 variant of COL6A3 to its Variant Panel. These tests are now available to horse owners in the EU and UK. The launch followed licensing of these tests from EquiSeq. Generatio GmbH – Center for Animal Genetics (CAG) has the exclusive rights to these tests in the EU and UK. For horses already tested for P2 (MYOT), P3 (FLNC), P4 (MYOZ3), and Px (CACNA2D3), a test for P8 (PYROXD1) and K1 (COL6A3) is available. Horse owners in the EU and UK can order the 6-Variant Panel online from the Animal Trust Center (ATC).
Blog
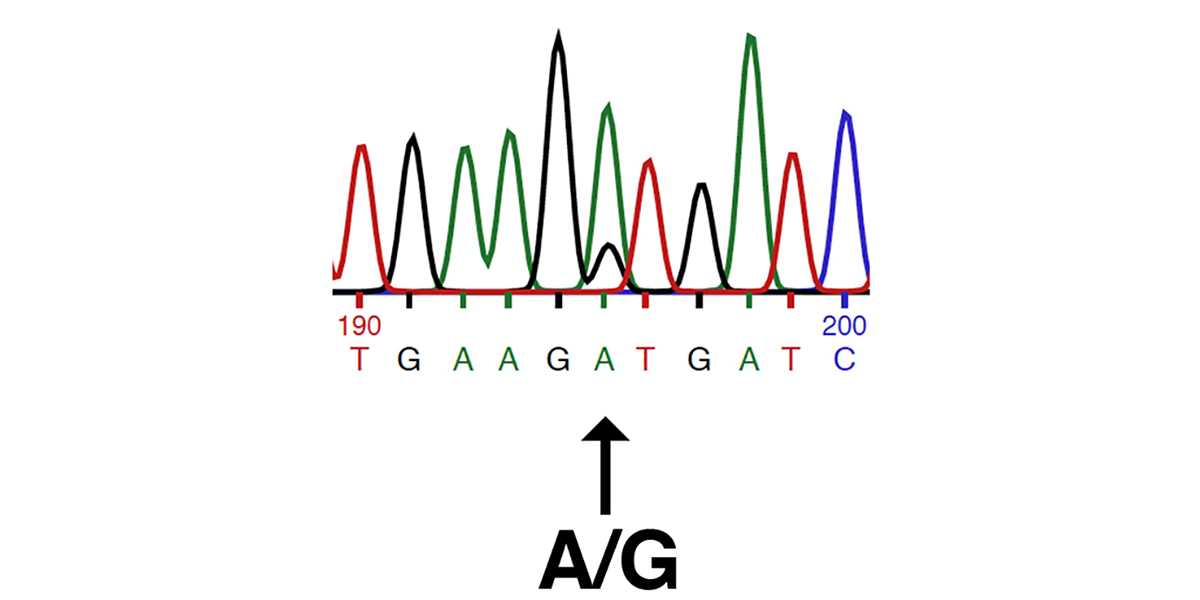
The K1 Genetic Variant Affects COL6A3, a Gene Encoding a Collagen
Contents Introduction The K1 genetic variant that has been part of EquiSeq’s Myopathy Panel since October 2019 is a missense allele of COL6A3, a gene encoding a collagen [1]. Collagens are a family of proteins that are the main structural components of the connective tissue. Collagens also guide bone formation and play an important role in many other tissues. They are the most abundant proteins in the body of mammals. There are thirty types of collagen, made up of collagen proteins encoded by 44 different genes. In humans, mutations in different collagen genes are responsible for a wide variety of inherited disorders. The phenotype produced by mutations in different collagens gives us an insight into the role of each of these collagens. The COL6A3 gene encodes one of the type VI collagens that are components of the endomysium, a layer of connective tissue that covers individual muscle fibers. Mutations in human COL6A1, COL6A2, and COL6A3 are associated with Bethlem myopathy and Ullrich congenital muscular dystrophy [1-11]. The K1 allele of equine COL6A3 is defined by its coordinates in the current genome assembly for horse, chr6:23,416,882 C/G in EquCab3.0 [12]. For purposes of this blog post, we will use the protein model XP_023498413.1, which would make this allele G2178A, the substitution…
Blog Uncategorized
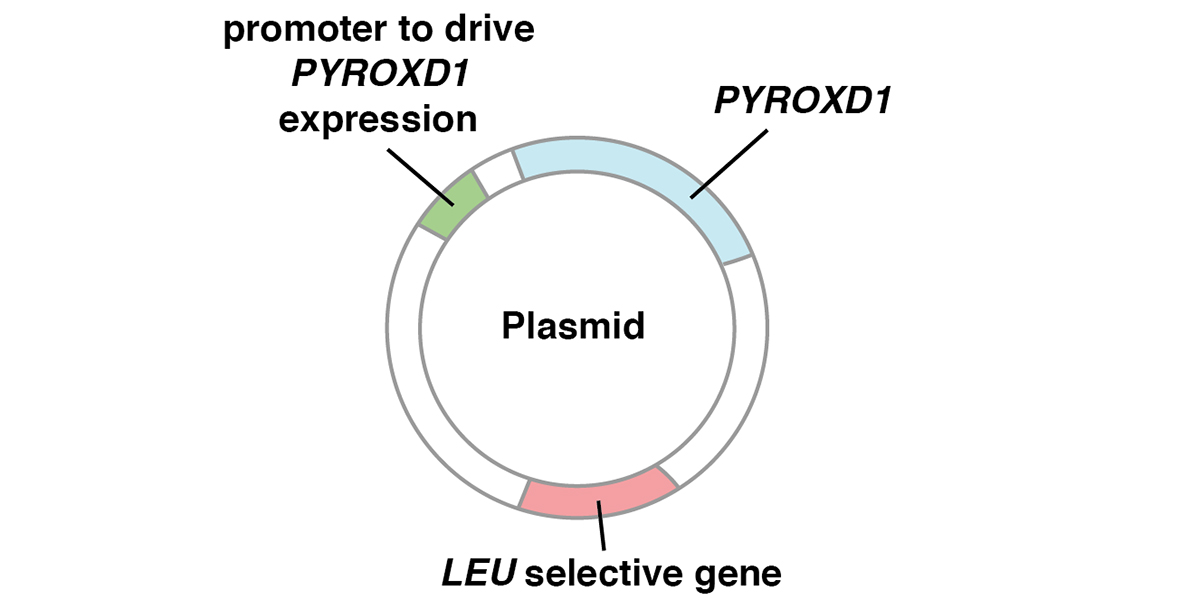
The P8 Genetic Variant Affects PYROXD1, a Gene Required for Oxidative Defense
Contents Introduction The P8 genetic variant that has been part of EquiSeq’s Myopathy Panel since October 2019 is a missense allele of PYROXD1, a gene required for oxidative defense [1]. Mutations of the human PYROXD1 gene are associated with Myofibrillar Myopathy 8 in humans [1]. The P8 allele of equine PYROXD1 changes an aspartic acid (D) to a histidine (H), a nonconservative substitution. Comparison of the amino acid sequence of the PYROXD1 protein across 248 species of vertebrates shows that the position affected by the P8 allele of PYROXD1 is highly conserved across hundreds of millions of years of evolution, with only aspartic acid (D) or asparagine (N) seen in this position. Preliminary biochemical evidence suggests that the P8 allele of PYROXD1 reduces the stability of the protein. There is independent evidence that the basis of Arabian MFM is a defect in oxidative defense affecting the redox state of cysteine thiols. Function of PYROXD1 PYROXD1 encodes pyridine nucleotide-disulfide oxidoreductase domain 1, an FAD-binding oxidoreductase that uses NAD/NADH as a cofactor to reduce cysteine thiols as part of the cellular oxidative defense. Mutations in the human PYROXD1 gene are associated with Myofibrillar Myopathy 8 (MFM8), a disease state defined by a breakdown of the integrity of the sarcomeric Z disc and myofibrils, accompanied by…
News
Podcast and Videos Explore Muscle Disease
Albuquerque, New Mexico July 2020 was a great month for horse owners seeking to understand muscle disease. Eden River Equestrian interviewed Paul Szauter, EquiSeq’s Chief Scientific Officer, for episode 86 of their Come Along for the Ride podcast. In the interview, Dr. Szauter talked about the effects of the genetic variants that are included in EquiSeq’s Myopathy Panel, with an emphasis on symptoms that will be evident to horse owners. He specifically recommended a video on PSSM & MFM Symptoms at canter by Christine Lola Mahon of the PSSM & MFM Awareness Group on Facebook. After the podcast was aired, Christine Lola Mahon posted a detailed video showing the progression of PSSM2/MFM, The Harsh Reality of PSSM2. The Equine Extension Program at the University of Minnesota hosted a set of four seminars by Dr. Molly McCue of the Equine Genetics and Genomics Laboratory. The seminars were held as Facebook Live events. Horse owners watching in real time were able to post questions and comments. The seminars are available as Facebook videos as listed below. Part 1 Introduction to Muscle Disease Part 2 PSSM Part 3 RER & MFM Part 4 HYPP & IMM Dr. McCue encouraged horse owners to participate in the University of Minnesota muscle disease study. Please see their FAQs or contact…
Blog
Peer Review Fails to Identify Errors in Inconclusive Study of Genetic Variants of Equine Muscle Genes
This blog post is a discussion of a paper published by Williams et al. in a recent issue of Equine Veterinary Journal [1]. The paper aims to identify genetic variants that might be responsible for Myofibrillar Myopathy (MFM) in Warmbloods. The authors use a candidate gene approach, searching for variants of eight of the nine genes known to be associated with MFM in human patients and eight additional genes. Variants are identified by RNA-Seq of 16 warmbloods typed by muscle biopsy (8 affected and 8 control), and by analysis of additional whole genome sequence data available to the authors and in public databases. The citation is: Zoe J. Williams (will3084@msu.edu), Deborah Velez-Irizarry (no contact information), Jessica L. Petersen (jessica.petersen@unl.edu), Julien Ochala (julien.ochala@kcl.ac.uk), Carrie J. Finno (cjfinno@ucdavis.edu) and Stephanie J. Valberg (valbergs@cvm.msu.edu) (2020) Candidate gene expression and coding sequence variants in Warmblood horses with myofibrillar myopathy. Equine Vet J. 00: 1-10. PMID: 32453872. Contents Summary Ninety variants were identified in exons of thirteen of the sixteen genes (ACTA1, BAG3, DES, DNAJB6, FLNC, HSPB8, KY, LDB3, LMNA, MYOT, PLEC, PYROXD1, and SQSTM1). No coding variants were found for CRYAB, FHL1, or TIA1. Of the 90 variants, 62 were synonymous substitutions, 2 were splice region variants, and 26 were missense alleles. All 26 missense alleles (in 11 genes) were predicted to have moderate effects…
Blog
Genetic Basis of Exercise Intolerance in Arabians
Paul Szauter, PhD, Chief Scientific Officer of EquiSeq, gave a presentation titled “Genetic Basis of Exercise Intolerance in Arabians” at the Al Khamsa Annual Meeting and Convention in Fayetteville, Arkansas on October 13, 2019. EquiSeq is a company based in Albuquerque, New Mexico, that develops and sells genetic tests for horses. Exercise intolerance is known in Arabians (1, 2) and limits the performance of some affected horses in endurance events. Symptoms of exercise intolerance (tying up) include sudden lameness, myoglobinuria, and greatly increased serum levels of creatine kinase (CK), a muscle enzyme released into the bloodstream when muscles are damaged. Researchers from EquiSeq began studying exercise intolerance in Arabians at an endurance event at the Biltmore estate in 2017. They were stationed at the vet trailer in order to study horses pulled from the event for symptoms of lameness. The weather was unusually cold and rainy, and it wasn’t long before the first horse showed up at the vet trailer with urine the color of black coffee and obvious symptoms of pain and lameness. A test for serum CK (normally in the range of 500 or less) showed levels in excess of 20,000. Eight hours and thirty liters of IV…
News
EquiSeq’s Kirsten Dimmler Accepted to PhD Program at the University of Minnesota
Albuquerque, NM Kirsten Dimmler, a Bioinformatics Analyst at EquiSeq, has accepted an offer of admission to graduate school in the College of Veterinary Medicine at the University of Minnesota. Kirsten will be entering the laboratory of Dr. Molly McCue, a leading veterinary expert in the field of equine genetics and genomics. Her admission to graduate school was originally scheduled for the fall of 2020, but has been deferred to January 2021 due to the COVID-19 pandemic. Kirsten began her career at EquiSeq with an internship while still an undergraduate biology major at the University of New Mexico. Following her graduation with a B.S. in Biology in December 2017, she began work at EquiSeq in bioinformatics. Her research involved evaluating potentially pathogenic mutations discovered in whole genome sequence data from horses with symptoms of exercise intolerance. The work led to a patent filing and the development of commercial genetic tests for horses. Kirsten was born and raised in Albuquerque, New Mexico. She has a passion for animals and had a goal of one day becoming a veterinarian. She purchased her first horse, an Appendix Quarter Horse mare named Esperanza, as a teenager and went on to compete in hunter/jumper with her….
Blog
The Px Allele of CACNA2D3 May Affect Splicing of a Component of the Voltage-Gated Calcium Channel
A Genome Wide Association Study (GWAS) of Recurrent Exertional Rhabdomyolysis (RER) identified a region on chromosome 16 as the only part of the genome associated with RER [1]. One of the populations in the study showed an association between RER and five Single Nucleotide Polymorphisms (SNPs) that define a 71.6 kb interval within the 822 kb CACNA2D3 gene. The CACNA2D3 gene encodes a regulatory subunit of the voltage-gated calcium channel called the dihydropyridine receptor (DHPR) that functions in excitation-contraction coupling, the process through which signals from motor neurons initiate muscle contraction [2]. The alpha(2)delta subunit encoded by CACNA2D3 sets the abundance of voltage-gated calcium channels by acting as a molecular chaperone. The alpha(2)delta subunit is bound to the plasma membrane through a glycosylphosphatidylinositol (GPI) anchor [3,4]. The level of expression of alpha(2)delta subunits sets the abundance of voltage-gated calcium channels in both neuronal and transfected nonneuronal cells [5,6]. The dihydropyridine receptor, of which the CACNA2D3 protein is a part, is known to physically interact with the ryanodine receptor encoded by the RYR1 gene. Malignant Hyperthermia (MH) in horses is caused by mutations in the RYR1 gene [7]. The RYR1-R2454G allele that causes MH in horses acts as an enhancer of the GYS1-R309H allele of glycogen synthase…
Blog
Mechanism of muscle damage in filaminopathy
Several recent studies address the mechanism of muscle damage in filaminopathy. Filaminopathy is a term that describes muscle disorders resulting from mutations in the FLNC gene, which encodes filamen C. Two different clinical disease states are included in filaminopathy: Myofibrillar Myopathy 5 (MFM5) and Distal Myopathy 4 (DM4). Studies on the mechanism of muscle damage in filaminopathy suggest that there is an overlap of symptoms between these two disease states, and that the same mechanism of muscle damage takes place in both. Muscle tissue from patients with MFM5 shows cytoplasmic protein aggregates and rimmed vacuoles; electron microscopy shows Z disc fragmentation, Z disc streaming, nemaline rods, and autophagic vacuoles. Muscle tissue from patients with DM4 shows nonspecific changes, not including the Z disc fragmentation and protein aggregates seen in MFM5. Studies of Human Patients A study of muscle tissue from human filaminopathy patients with the W2710X or V930-T933del mutations shows that the large protein aggregates that are seen in Myofibrillar Myopathy contain not only Z disc proteins, but also proteins involved in clearing protein aggregates [1]. Double immunofluorescence with antibodies to either FLNC or MYOT and a series of other proteins shows that the aggregates contain heat shock proteins (HSP20, HSP27, HSP40,…